GoodScents and Leffingwell Molecular Odours
Data Science and Analytics
Tags and Keywords
Trusted By



"No reviews yet"
Free
About
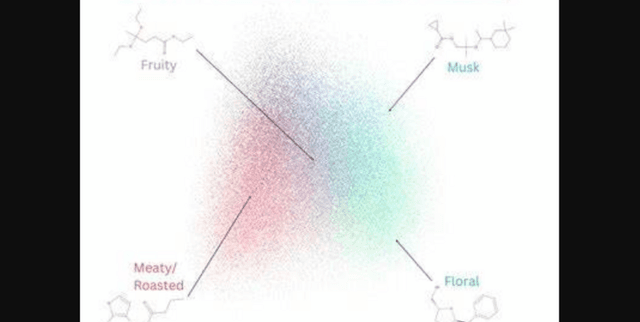
Multi-labelled chemical structure collection comprising 4,983 molecules associated with 138 distinct odour descriptors. Constructed to facilitate the replication of the "Principal Odor Map" research, this resource aggregates data from GoodScents and Leffingwell PMP 2001. It supports tasks in olfactory perception, graph neural networks, and multi-label classification by linking molecular structures (SMILES) with perceptual labels such as 'fruity', 'woody', and 'creamy'.
Columns
- nonStereoSMILES: The molecular structure represented in Simplified Molecular Input Line Entry System (SMILES) format.
- descriptors: A text string concatenating all applicable odour labels for the molecule (e.g., "odorless", "fruity").
- alcoholic: Binary indicator (0 or 1) identifying if the molecule possesses an alcoholic scent.
- aldehydic: Binary indicator for an aldehydic scent.
- alliaceous: Binary indicator for an onion or garlic-like scent.
- almond: Binary indicator for an almond scent.
- amber: Binary indicator for an amber scent.
- animal: Binary indicator for an animalic scent.
- anisic: Binary indicator for an anise scent.
- apple: Binary indicator for an apple scent.
- [Additional Label Columns]: The dataset includes further binary columns for the remaining 130 descriptors, covering scents such as banana, beefy, citrus, creamy, fishy, floral, grassy, musky, spicy, vanilla, and woody.
Distribution
- Format: CSV (Multi-Labelled_Smiles_Odors_dataset.csv)
- Size: 1.65 MB
- Rows: 4,983 unique records
- Structure: 140 columns (comprising the SMILES structure, descriptor string, and 138 specific binary odour labels).
- Data Quality: 100% valid entries with 0% missing values across the documented columns.
Usage
- Training Graph Neural Networks (GNN) for molecular property prediction.
- Developing multi-label classification models for olfactory perception.
- Replicating the findings of the "Principal Odor Map" study.
- Analysing Structure-Odour Relationships (SOR) in chemoinformatics.
- Benchmarking machine learning models in biotechnology and chemistry.
Coverage
- Geographic/Source Scope: Aggregated from the GoodScents and Leffingwell PMP 2001 databases.
- Subject Scope: Covers 4,983 specific odorant molecules.
- Descriptor Scope: Spans 138 perceptual categories ranging from food-related notes (chocolate, coffee) to chemical (ethereal, solvent) and natural tones (earthy, mossy).
License
CC0: Public Domain
Who Can Use It
- Computational Chemists
- Machine Learning Engineers specialising in GNNs
- Olfactory Researchers
- Flavour and Fragrance Scientists
- Biotechnology Students
Dataset Name Suggestions
- SMILES Odor Descriptors for Principal Odor Map
- Multi-Label Olfactory Perception Database
- GoodScents and Leffingwell Molecular Odours
- Chemical Structure and Odour Label Collection
Attributes
Original Data Source: GoodScents and Leffingwell Molecular Odours
Loading...
