Raw Data Supporting Adenocarcinoma Prediction
Patient Health Records & Digital Health
Tags and Keywords
Trusted By



"No reviews yet"
Free
About
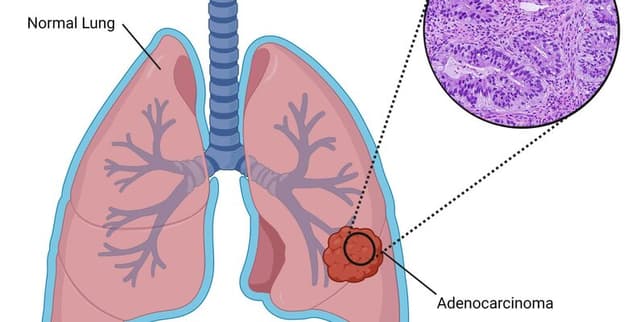
This collection contains all the raw data and analysis methodologies used for predicting Lung Adenocarcinoma based on specific cell-death features. The data is pivotal for validating the findings that combining multiple cell-death indicators offers significant predictive power in recognising this specific type of lung cancer. The dataset includes quantified gene expression levels alongside associated survival outcomes for study subjects.
Columns
The data structure includes several key columns:
- ID (e.g., GSM773540): Unique identifier for each sample or patient record.
- IDO1: Expression level data for the IDO1 gene.
- GJB2: Expression level data for the GJB2 gene.
- PTAFR: Expression level data for the PTAFR gene.
- BIRC3: Expression level data for the BIRC3 gene.
- status: An indicator variable representing the survival status or outcome.
- os: Overall survival time, measured in days.
Distribution
The raw data for the manuscript is organised across three directories. The collection includes 5 primary raw data files structured to support gene expression and survival analysis, along with 13 files containing R language code used for the analytical procedures. The expected frequency of updates for this material is never.
Usage
This data is ideal for applications in oncology research, particularly for researchers interested in lung cancer prediction and prognosis. It can be used to replicate the predictive models detailed in the associated manuscript, study the correlation between specific gene expressions (IDO1, GJB2, PTAFR, BIRC3) and survival outcomes, and develop new statistical models for biomarker validation in adenocarcinoma.
Coverage
The material focuses on biological measurements (gene expression and survival metrics) related to Lung Cancer. Specific details regarding the geographic location, time range of data collection, or patient demographics are not specified within the available details.
License
CC0: Public Domain
Who Can Use It
The primary intended users include academic researchers and clinical scientists focused on cancer biology and genomics. Data scientists can utilise the metrics for machine learning and statistical modelling tasks related to disease prediction, and computational biologists can use the gene expression profiles for functional enrichment analysis.
Dataset Name Suggestions
- Cell-Death Features Predictive Data for Lung Adenocarcinoma
- Gene Expression and Survival Metrics in LUAD
- Raw Data Supporting Adenocarcinoma Prediction
- IDO1 GJB2 PTAFR BIRC3 Gene Expression Lung Cancer
Attributes
Original Data Source: Raw Data Supporting Adenocarcinoma Prediction
Loading...
