Enriched Human Macromolecule Structure Dataset
Data Science and Analytics
Tags and Keywords
Trusted By



"No reviews yet"
Free
About
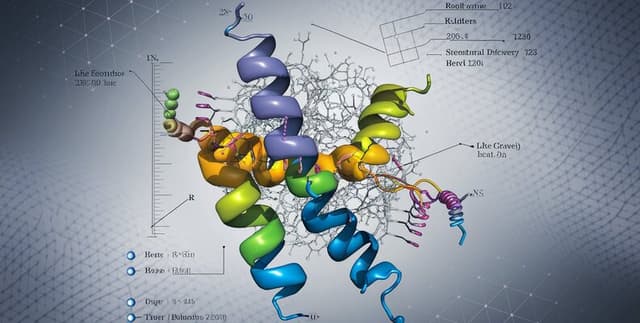
Contains 11,832 macromolecular structures of human proteins sourced from the RCSB Protein Data Bank (PDB). This dataset is enriched with new structural features, including the number of residues, chains, and secondary structure components like helices, sheets, and coils. The data covers proteins determined by X-ray crystallography between 2015 and 2023, filtered for high quality with resolutions from 1.0 Å to 3.0 Å. It is specifically curated to support various machine learning and deep learning applications in structural bioinformatics, such as protein-ligand binding prediction, enzyme classification, and structure quality assessment.
Columns
- PDB ID: Unique identifier for each protein structure.
- Experimental Method: The technique used to determine the structure, primarily X-ray diffraction.
- Matthews Coefficient: An estimate of the crystal's volume occupied by protein versus solvent.
- Percent Solvent Content: Percentage of solvent within the crystal.
- Crystallization Method: The technique used for crystal growth, such as vapor diffusion.
- pH: The pH level at which crystallization occurred.
- Crystal Growth Procedure: A detailed description of the crystal growth process.
- Temp (K): Crystallization temperature in Kelvin.
- Deposition Date: The date the structure was submitted to the PDB.
- Release Date: The date the structure was made public.
- Number of Non-Hydrogen Atoms per Deposited Model: Count of non-hydrogen atoms in the structure.
- Total Number of Polymer Instances (Chains): The number of unique polymer chains.
- Total Number of Polymer Residues per Deposited Model: Total amino acid residues in the model.
- Number of Water Molecules per Deposited Model: Total water molecules in the structure.
- Disulfide Bond Count per Deposited Model: The number of disulfide bonds.
- Molecular Weight per Deposited Model: Molecular weight of the entire structure.
- Number of Distinct Protein Entities: The count of unique protein entities.
- Refinement Resolution (Å): Resolution of the X-ray experiment in Ångströms.
- Structure Determination Methodology: The overall method used, e.g., 'experimental'.
- Average B Factor: A measure of atomic displacement, indicating flexibility.
- R Free: A validation metric for the crystallographic model quality.
- R Work: A measure of how well the model fits experimental data.
- Structure Title: The title assigned by the researchers.
- Sequence: The amino acid sequence of the protein.
- Entity Polymer Type: The type of polymer, e.g., 'Protein'.
- Polymer Entity Sequence Length: The length of the polymer sequence.
- Entity Macromolecule Type: The type of macromolecule, e.g., 'polypeptide(L)'.
- Total Number of Polymer Entity Instances (Chains) per Entity: Number of chains per polymer entity.
- Molecular Weight (Entity): Molecular weight of an individual polymer entity.
- Macromolecule Name: The name of the macromolecule.
- EC Number: The Enzyme Commission number for enzyme classification.
- EC Provenance Source: The source of the enzyme classification.
- Source Organism: The organism from which the protein was derived.
- Taxonomy ID: Identifier for the source organism's taxonomy.
- Total Number of Polymer Residues per Assembly: Total residues in the full assembly.
- Total Number of Polymer Instances (Chains) per Assembly: Total chains in the full assembly.
- Oligomeric Count: The number of subunits in the oligomeric state.
- Assembly ID: A unique identifier for the assembly.
- Oligomeric State: The functional form of the protein (e.g., 'Monomer').
- Stoichiometry: The ratio of components in the protein assembly.
- Ligand ID: Identifier for any bound ligand.
- Ligand Formula: The chemical formula of the ligand.
- Ligand MW: The molecular weight of the ligand.
- Ligand Name: The common name of the ligand.
- InChI: The International Chemical Identifier for the ligand.
- Ligand of Interest: Indicates if a ligand is of special interest.
- Number of Residues: Total count of amino acid residues (newly added).
- Number of Chains: Count of distinct chains in the structure (newly added).
- Helix Count: Number of alpha-helices (newly added).
- Sheet Count: Number of beta-sheets (newly added).
- Coil Count: Number of random coil regions (newly added).
Distribution
- Format: A single CSV file named
RCSB_PDB_Macromolecular_Structure_Dataset.csv. - Size: 11.36 MB.
- Structure: The dataset is tabular and contains 11,832 records (rows) and 46 columns.
Usage
This dataset is optimized for a variety of machine learning and deep learning tasks, including:
- Protein-ligand binding prediction.
- Oligomeric state analysis and prediction.
- Enzyme classification using EC numbers.
- Protein secondary structure prediction.
- Assessment of protein structure quality.
- Protein stability and protein-protein interaction prediction.
- Protein domain analysis and evolutionary analysis.
- Prediction of optimal protein crystallization conditions.
Coverage
- Geographic: Not applicable, as the data is molecular. The source organism is predominantly Homo sapiens (human).
- Time Range: The dataset includes protein structures with release dates from January 2015 to September 2023.
- Demographic: The data pertains to human (Homo sapiens) proteins.
License
CC0: Public Domain
Who Can Use It
- Bioinformaticians and Computational Biologists: For research in structural biology, protein function prediction, and evolutionary analysis.
- Data Scientists and Machine Learning Engineers: For developing and training models on tasks like enzyme classification, ligand binding prediction, and protein stability analysis.
- Pharmaceutical Researchers: For drug discovery, identifying potential binding sites, and analysing protein-ligand interactions.
- Academics and Students: For educational purposes and research projects in biochemistry, molecular biology, and data science.
Dataset Name Suggestions
- Human Protein Structures for Machine Learning
- RCSB PDB Human Protein Structural Features
- Enriched Human Macromolecule Structure Dataset
- ML-Ready Human Protein Data (2015-2023)
- High-Resolution Human Protein Structures with Ligands
Attributes
Original Data Source: Enriched Human Macromolecule Structure Dataset
Loading...
Free
Download Dataset in ZIP Format
Recommended Datasets
Loading recommendations...
