Bio-Inspired Iris Temporal Encoding Dataset
Data Science and Analytics
Tags and Keywords
Trusted By



"No reviews yet"
Free
About
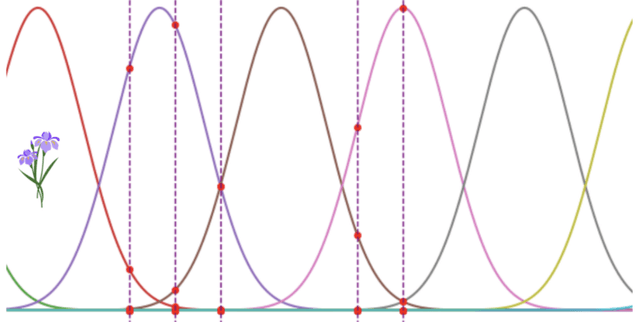
Encoding the classic Iris dataset through Gaussian receptive fields facilitates its application within spiking neural networks (SNN). By transforming quantitative features—sepal and petal dimensions—into temporal spike latencies, this resource allows for the exploration of bio-inspired computing models without relying on pre-built SNN libraries. The resulting information represents the excitation levels and spike timings of 40 presynaptic neurons, providing a bridge between traditional tabular measurements and the temporal coding required for advanced neural simulations.
Columns
- № presynaptic neuron: The unique identifier for each of the 40 neurons involved in the encoding process, with 10 neurons assigned to each of the four physical features.
- 0-149: Individual columns representing the encoded spike latencies or excitation values for each of the 150 samples from the original floral collection.
Distribution
The information is delivered in a CSV file titled
Iris_Dataset_encoded_by_Gaussian_receptive_fields_for_SNN.csv with a file size of 22.01 kB. It contains 40 records representing the presynaptic neurons, structured across 151 columns. The resource maintains a usability score of 10.00 with 100% validity and no mismatched or missing values in the primary neural indices.Usage
This resource is designed for training and testing spiking neural networks, particularly those focused on temporal encoding and Spike-Timing-Dependent Plasticity (STDP). Researchers can use it to benchmark SNN reinforcement learning methods or to study unsupervised clustering with spiking neurons. It is also an excellent tool for demonstrating Gaussian receptive field encoding techniques within the field of computational neuroscience.
Coverage
The scope encompasses the three classic Iris flower species: Setosa, Versicolor, and Virginica. It covers four physical attributes—sepal length, sepal width, petal length, and petal width—transformed into 40 distinct neural signals. The collection provides a static representation of the original 150 observations processed into the temporal domain.
License
CC0: Public Domain
Who Can Use It
Computational neuroscientists can apply these encoded signals to test the efficiency of temporal coding in spiking models. Machine learning engineers interested in bio-inspired AI can use the pre-processed latencies to bypass the encoding stage of their simulations. Additionally, academic students studying neural networks can use the records to understand the mathematical transition from static data to spike-based information representation.
Dataset Name Suggestions
- Gaussian Encoded Iris Dataset for Spiking Neural Networks
- Iris Species Temporal Spike Latency Registry
- Gaussian Receptive Field Encoded Iris Data
- SNN-Ready Iris Feature Latency Index
- Bio-Inspired Iris Temporal Encoding Dataset
Attributes
Original Data Source: Bio-Inspired Iris Temporal Encoding Dataset
Loading...
Free
Download Dataset in CSV Format
Recommended Datasets
Loading recommendations...
