Refined Neuronal Cell Instance Segmentation Labels
DNA & Genomics Data
Tags and Keywords
Trusted By



"No reviews yet"
Free
About
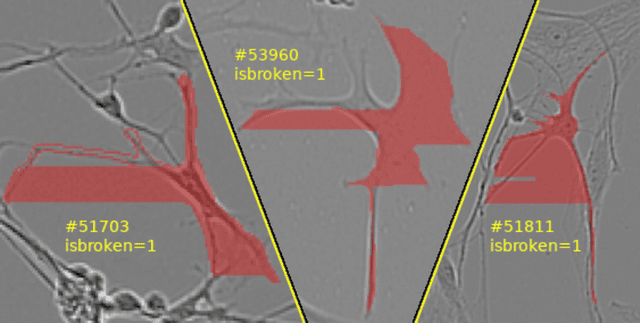
Addressing the inaccuracies found in biological image annotations, this resource introduces a manual verification layer to the Sartorius Cell Instance Segmentation collection. It specifically targets the issue of "broken" labels—segments that may be undesirable for training high-precision models. By providing a clear indication of label integrity, the data allows for more refined training processes where problematic masks can be excluded without the need to discard entire images or associated valid labels. This effort represents three days of manual visual inspection to identify bad labels that could otherwise introduce noise into data science models.
Columns
- id: A unique identifier assigned to each biological object in the dataset.
- annotation: The run-length encoded pixel data representing the identified neuronal cell.
- width: The width of the source image, consistently recorded as 704 pixels.
- height: The height of the source image, consistently recorded as 520 pixels.
- cell_type: The specific cell line categorisation, such as shsy5y or cort.
- plate_time: The timestamp indicating when the plate containing the cells was created.
- sample_date: The date on which the biological sample was officially recorded.
- sample_id: A unique identifier for the specific sample being observed.
- elapsed_timedelta: The time elapsed since the initial image of the sample was captured.
- isbroken: A binary flag where 1 indicates the original label is broken and 0 indicates it is valid.
Distribution
The dataset is provided in a single CSV file named
train.csv with a total file size of 24.61 MB. It contains 73.6k valid records across 10 distinct columns. The data structure is highly consistent, with 100% validity for identifiers, dimensions, and the refined "isbroken" flag. It is provided as a static release with no further updates expected.Usage
This resource is ideal for improving the performance of instance segmentation models by filtering out low-quality or corrupted training masks. It is well-suited for researchers who wish to perform label refinement or study the impact of noisy labels on deep learning outcomes in medical imaging. Additionally, it can be used to benchmark how effectively various models handle real-world data containing human-identified errors.
Coverage
The geographic and demographic scope is focused on biological cell lines (specifically shsy5y and cort) within a laboratory environment. Temporally, the data captures observations from 14 June 2019 through to 7 November 2020. While the "isbroken" flag was generated through careful manual inspection, users should note the possibility of minor human error in the flagging process.
License
CC BY-NC-SA 4.0
Who Can Use It
Data scientists can leverage these records to clean their training pipelines for the Sartorius competition or similar segmentation tasks. Computational biologists may utilise the refined labels to ensure that their automated cell counts and boundary detections are based on accurate ground truths. Furthermore, machine learning researchers can use this as a case study for "broken" mask management in computer vision.
Dataset Name Suggestions
- Sartorius Cell Segmentation: Broken Label Refinement Index
- Refined Neuronal Cell Instance Segmentation Labels
- Biological Image Label Quality Index: The "isbroken" Flag
- Sartorius Training Data: Manual Label Integrity Registry
- Cleaned Instance Segmentation Masks for Neuronal Cells
Attributes
Original Data Source: Refined Neuronal Cell Instance Segmentation Labels
Loading...
Free
Download Dataset in CSV Format
Recommended Datasets
Loading recommendations...
